The case for ancient tree genetics
We’re arguing that the combination of government policy for widespread woodland creation and modern tree production processes are potentially narrowing the scope of genetic variability within the UK’s future treescape. So isn’t it strange that we’re advocating the use of a small number of ancient trees to act as seed sources? Here’s why we don’t think this is a problem.

Start with thinking about the genetic variation that resides within a whole population, say all the English oaks in the country. If you were to sample that entire population you’d find out how much the species varies across its UK range.
If you went to a more local population, such as within one of the Forestry Commission’s seed sourcing zones, and sampled every tree you’d find a subset of the total variability.
Sampling any individual tree would unsurprisingly yield a further subset of genetic variability. In simple terms the smaller the sample the less proportion of the total genetic variability is captured.
It turns out that for many tree species, that first subset is likely to capture a significant proportion of the total population’s genetic variability. A recent study at Wytham Woods in Oxfordshire showed that there is more genetic variation within populations that between populations.
However, there are also signs of inbreeding within younger populations of trees. That could be a result of the increased reliance on a relatively small number of seed sources, which supply the commercial tree nursery trade. This practice could be resulting in a narrowing of genetic diversity, albeit without deliberate selection for particular traits.

Where ancient trees fit in
Ancient trees represent a genetic time capsule. Their genomes are a subset of the population at the time they germinated. The population as a whole will have changed over time due to a combination of genetic drift, natural selection and artificial selection. The latter might be deliberate, such as through selection for particularly favourable traits, or inadvertent, which might be through the use of a convenient, productive seed source.
The effect is illustrated below. Any given trait will have a range of values, which in this example are normally distributed about a mean. An individual tree at time T=0 is denoted by the red dot. In this case its trait value is close to the population mean.
Over the next hundred years, evolutionary forces cause the population mean to shift slightly to the left. Our tree is now something of a rarity, being more than one standard deviation (hatched area) away from the mean.
A hundred years later, at T=200, selection has moved the mean back nearly to where it started when our tree germinated. Once again it’s just an average tree.
But after 400 years, when our tree is ancient among its species, its trait value lies well outside the normal range. Quite possibly, such values are being selected against and could be deleterious. But much depends on factors such as when in the tree’s life history the trait becomes relevant, whether it is linked to other traits, and the individual tree’s capacity to survive despite having this particular trait value.
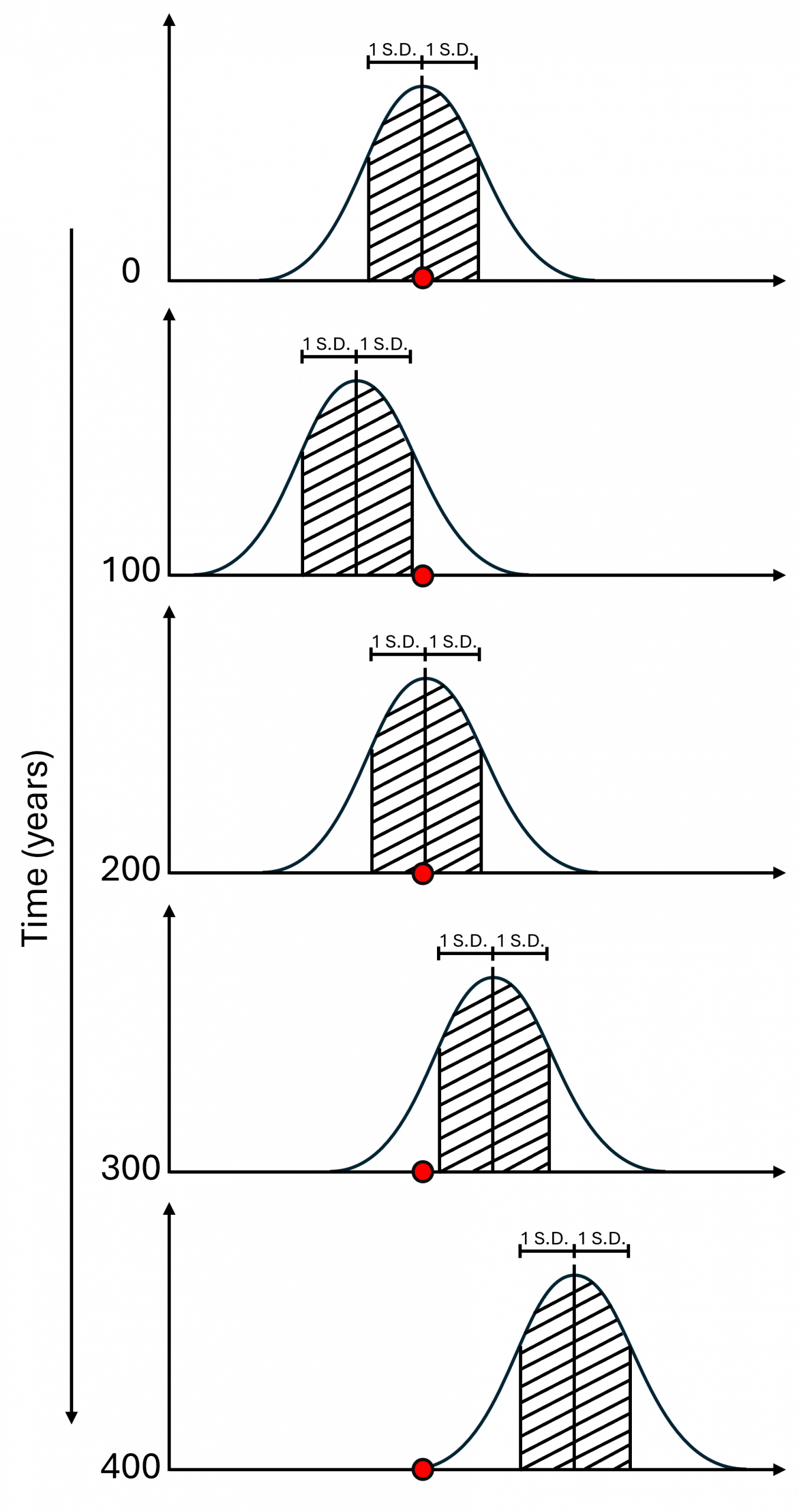
The illustration above is greatly simplified but hopefully makes the point about why ancient trees are important from a genetic perspective. They have an unusually high chance of possessing trait values that lie outside the normal range for the population, so the inclusion of their progeny in contemporary woodland creation increases the range of genetic diversity and potentially the population’s adaptive capacity.
